Genetic Methods of Species Identification
- Introduction
- Genetic IDs
- Why Evolution Matters
- Genomes
- Gene Regions Used in Species Identification
- Sequence Alignment
- Methods Used
- Using Phylogenetic Trees
- DNA Barcoding
- BLAST
- Are these Methods Reliable?
Introduction
When confronted with an unknown specimen there are lots of different ways of determining what species it is. We could use each of the following methods:
- appearance and visible traits - consider shape, colour, unique features
- microscopic analysis - search for types of cells, or cell structures
- chemical analysis - search for unique or diagnostic chemical compounds
- genetic analysis - search for unique or diagnostic genetic attributes
Which method we choose will depend on many factors. Sometimes the simplest approach, just looking at the specimen and comparing it with reference books, is not enough.
- In what state or form is the specimen? Do we have the whole organism or most of it, or are significant parts missing? For example, is the animal all there, or do we just have a piece of tissue, such as meat in the freezer? Perhaps all we have is a blood stain or tuft of feathers.
- Is the specimen from a group of well-studied species and do we know how to distinguish them one from another? Some species look the same at certain stages of their life cycle.
Here we will consider how genetic analysis can be used to identify specimens.
Genetic Analysis
Nowadays, genetic analysis involves some form of DNA sequencing. That means the sequence of A, C, G and T nucleotides is determined for a specific region of the DNA extracted from the specimen. This sequence can then either
- be examined for genetic traits which are characteristic or diagnostic for a particular species, or
- be used to investigate its evolutionary relationship with known species
Why Evolution Matters
Genetic analyses are successful because genetic differences accumulate slowly as species evolve. For any particular part of a species' genome, genetic changes accumulate as time passes. When that species becomes split into two species then each of those daughter species will accumulate different changes. They will however share the changes which their ancestor acquired. So, species contain both unique differences, which might be diagnostic, and shared differences which might be used to study relationships with other species.
Genomes
All plants and animals have a large genome in the nucleus of (most of) their cells. This genome is usually seen in the form of chromosomes. These organisms also have other genomes. Both plants and animals have energy-producing cell structures called mitochondria which contain very small genomes. Plants have additional structures called chloroplasts, where photosynthesis occurs, which also have small genomes.
Gene Regions Used in Species Identification
Scientists interested in developing methods of species identification choose to study specific gene regions for very practical reasons. Their primary concerns are:
- Is there sufficient genetic material in the specimen for me to analyze?
- Is there enough genetic difference among species for me to be able to distinguish among them?
- Is the genetic difference among species small enough that it makes some sense?
They are generally not interested in whether the gene region is involved in some significant metabolic function or cellular process. In fact they might try to avoid using such regions. Why do you think that might be?
In general, they choose gene regions in the mitochondrial genome. There are thousands of copies of a mitochondrial gene per cell, rather than just the two copies of any gene in the nuclear genome. There are different rates of change in different gene regions. Fast-evolving regions such as the "D-loop" might be used for studying closely related species while slowly-evolving regions such as cytochrome b (CYTB) and cytochrome oxidase I (COX1) might be used for distantly related species.
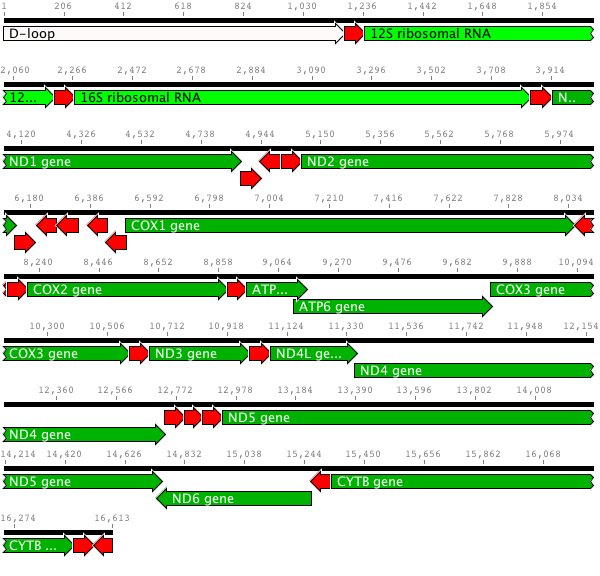
Here is the mitochondrial genome for the pig (Sus scrofa). Dark green represents genes which encode a protein, light green are ribosomal RNA, red are transfer RNA, and white represents other structures.
Note: Genetic analysis to identify specific individuals, say in human forensics or parent analysis, is based on a very different approach using genes from the nuclear genome.
Sequence Alignment
In many of the methods described here, the first step is to create a sequence alignment. Sequences from different reference species, and the unknown specimen, are brought together.

Then gaps ('-') are inserted to bring the sequences into alignment. The hypothesis is that nucleotides in the same column are the same or different by reason of evolution. The gaps represent either an insertion of genetic material in some sequences, or a deletion of material in others. Because we often don't know which happened, these are called indels.
Among the most commonly used methods of species identification are:
- Phylogenetic trees - estimate the evolutionary relationships among the unknown sequence and a set of known reference sequences
- DNA Barcoding - find the reference sequence from known species which is most similar to the unknown sequence
- BLAST - find the sequence in large public databases of genetic sequences which gives the best sequence alignment with the unknown sequence
© The University of Auckland 2020 | disclaimer
